VarioML Core Ontology
Specification
June 2011
This version:
https://svn.gene.le.ac.uk/gen2phen/trunk/data_formats/xml/html/LSDB_spec.html Latest version:
https://svn.gene.le.ac.uk/gen2phen/trunk/data_formats/xml/html/LSDB_spec.html Last update:
Date: 06/06/2011
Revision:
Revision: 1.0
Contact:
Juha
Muilu - FIMM Editors:
Name - Gen2Phen Name - Institution Contributors:
See acknowledgements .
Copyright © 2007-2011 by Gen2Phen
Development of VarioML Data format has been supported by the
European Community's Seventh Framework Programme (FP7/2007-2013)
under grant agreement number 200754 - the GEN2PHEN project
This work is licensed under the GNU GPL .
The VarioML Core Ontology provides the main concepts and properties
required to describe variant information used in clinical and research
databases. This document contains a detailed description of the
VarioML Core Ontology.
NOTE: This section describes the status of
this document at the time of its publication. Other documents may
supersede this document.
This specification is an evolving document. This document is
generated by combining a machine-readable
VarioML Core Ontology Namespace expressed in RelaxNG-compact
with a
specification template and a set of per-term documents.
The authors welcome suggestions on the
VarioML Core Ontology Namespace and this document. Please send
comments to the Gen2Phen
Web
Services & Exchange Formats group . Public
archives are available. This document may be updated or added
to based on implementation experience, but no commitment is made by
the authors regarding future updates.
Introduction
Background Standards Overview VarioML Examples VarioML Extensibility: Variant Report VarioML classes
and
properties cross-reference
Glossary External Classes and Properties Acknowledgements References Change
log
The VarioML data format supports data exchange between locus
specific databases (LSDBs) and central repositories, in the context of high-throughput (HTP) bioinformatic data flows.
Use cases include exchanging data between partners in the context
of specific phenotype extensions. External invited participants
from the epidemiology, medical genetics, ontology development and
model organism communities provided expertise and use cases beyond
those of Gen2Phen Partners.
Validation of this LSBD data model commenced in 2009 by working
with the existing LSDBs inside and outside the GEN2PHEN consortium,
most of whom have existing data formats. This
document describes XML
and MAGE-TAB based data
formats developed from the data model. The VarioML format complements the format developed for the Locus Reference
Genomic sequence (LRG) by providing
detailed descriptions for variant and individual (patient, or
other source of genetic data) information, as
defined in Gen2Phen workshops and subsequent community consultations.
The MAGE-TAB format is meant for simple
spreadsheet-based data
entry, covering a core set of VarioML information defined
in XML.
The complex integration of data from patients to
alleles is organized and standardized, using
groups to further simplify research that uses and adds to the growing
resources available over the net.
Abstract classes are used to make the VarioML format compatible with
other ontologies in use around the world.
Future plans include a tabular
"light" format, based on the
XML format described here, for use by genome browsers such as
Ensembl .
The VarioML format is managed as a collaborative effort among
members of the Gen2Phen
consortium , funded by the Health Thematic Area of the
Cooperation Programme of the European Commission within the VII
Framework Programme for Research and Technological Development. The
name "VarioML" is an acronym for "Locus Specific Database".
More resources on LSDBs are available at the Gen2Phen project site .
The Gen2Phen
Web Services and Exchange Formats
group is the main discussion list for questions about the
VarioML format.
The remainder of this specification describes how to publish and
interpret descriptions such as these on the Web, using XML for
syntax (file format) and terms from VarioML. It introduces a number of
classes (concepts such as variant and exon)
and properties (relationship
and attribute types such as DNA structure and transversion).
The specific contents of the VarioML Core Ontology are detailed in the VarioML
Core Ontology Namespace document .
This specification uses the XML format for ease of adoption, but
has been designed with awareness of the growing importance of VarioML
data becoming available in Linked Data/RDF form. This document has been
structured to easily be adaptable to RDFized VarioML data.
For more information, see:
[back to
top ]
Standards
The VarioML data format merges data models and expertise from
epidemiology, medical genetics, ontology development and model
organism communities. The most recent step in developing this
standard is the inclusion of Mauno Vihinen's Variation Ontology .
This document presents VarioML as a Semantic Web vocabulary or
ontology . It describes the VarioML Core Ontology and the
terms (XML classes and
properties) that constitute it, so that Semantic Web applications can
use
those terms in a variety of XML- and eventually RDF-compatible document
formats and
applications. The VarioML Core Ontology is straightforward, pragmatic
and designed to allow simultaneous deployment and extension, and is
therefore intended for wide scale use.
The VarioML ontology is identified by the
namespace URI:VarioML:namespace: http://gen2phen.org/VarioML/1.0
Note that ordering of XML classes is significant in
formats, e.g. to ensure accurate multiplicity relations. Properties can
occur in any order. See here for ordering of
classes. Class names are capitalized; property names are lower-case.
VarioML Schemata
RelaxNG
Compact schema definition language. Relax files can be used with any RelaxNGC validator
(currently Jing has been tested). Oxygen
tool, which uses the open
source Trang
schema
converter . The XML-schema can be downloaded here:
:
VarioML Validation
Validate XML made with the VarioML spec here
Validate RDF made with the VarioML spec here
Example: validation
of sample VarioML submission Example submission is here
[back to
top ]
VarioML Exchange Format
The VarioML core ontology is expressed as an XML schema using the RelaxNG-compact schema definition
language , following implementation patterns used
in the LRG XML format. This format is based on a minimum
information checklist being developed in Gen2Phen workshops, available here .
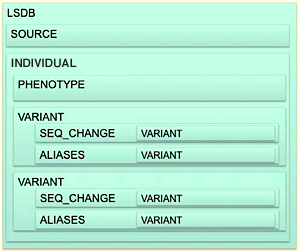
Figure 1: Overall description of the format, in an individual-centric implementation
Description of the format
Given an individual-centric use case, the overall structure is shown in Figure 1. The
format has a single root element LSDB: , with the following
sub elements:
The source element group provides data
source information such as database name, contact person(s), etc. There
can be multiple source elements, since data can originate from multiple
sites.
The individual element
describes the phenotype of the individual using optional ontology terms
and optional references to detailed
phenotype descriptions, which are defined outside the VarioML
format.
The variant element group specifies
mutation specific information,
from DNA to RNA and aminoacid levels, using variant elements
embedded recursively in the seq_changes
element .aliases element ,
specifying variants typically named using a different coordinate system
or naming
scheme.
VarioML has been designed to serve the spectrum of data exchange needs for variant data, which may be individual-, variant-, or population centric. Separate schema instantiate the VarioML core ontology seperate schema for these needs.
The LSDB-XML format is individual-centric, where the
individual can be a patient or other source of genetic data, for locus
specific
and clinical databases. However, there is also a need for VarioML data
exchange in a variant-centric format, for variant reports on
population frequency levels. The VREPORT-XML
module , described below, provides a simple adaption of the VarioML
format for this purpose.
[back to
top ]
<variant type="cDNA">
<lsdb id="submission1" schema_version="2.0" xmlns="http://gen2phen.org/varioml/2.0"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://gen2phen.org/lsdb/2.0 file:gen2phen/trunk/data_formats/xml/lsdb.xsd">
<source id="fimm">
<name>Insitute for Molecular Medicine Finland (FIMM)</name>
<contact>
<name>J. Muilu</name>
</contact>
</source>
<individual id="id1">
<gender code="1"/>
<variant id="id3">
<ref_seq accession="XYZ"></ref_seq>
<name scheme="HGVS"> c123C>G </name>
</variant>
<variant id="id5">
<ref_seq accession="XYZ"></ref_seq>
<name scheme="HGVS"> c123C>G </name>
</variant>
</individual>
<individual id="id7">
<gender code="9"><description term="XX-male"/></gender>
<variant id="id9">
<ref_seq accession="XYZ"></ref_seq>
<name scheme="HGVS">
c123C>G
</name>
</variant>
</individual>
<comment><text>simple comment</text></comment>
NB: Gender codes are 0:
'unknown', 1: 'male', 2: 'female' and 9: 'not applicable'.
[back to
top ]
A more detailed example of a VarioML submission:
<lsdb id="submission0001" xmlns="http://gen2phen.org/varioml/2.0"
[back to
top ]
VarioML has been designed to be extensible to serve the spectrum of variant data exchange needs. We have included here one such extension: a Variant Report format to serve the need for data
exchange in a variant-centric format. This also serves as an example of how to extend the VarioML core ontology for diverse use-cases.
VREPORT-XML
Variant Reporting makes use of a VREPORT-XML schema, extending the available VarioML terms to provide a
simple adaption for making variant reports on population frequency levels.
Where LSDB-XML is individual-centric, for locus specific and
clinical databases, VREPORT-XML is population-centric, using the Panel class, for variant
reports on population frequency levels.
VREPORT extends the VarioML model for use with National and Ethnic Mutation Databases (NEMDBs).
VREPORT-XML format differs from the LSDB-XML format in two major ways:
Extending VarioML with domain-specific Schema
The VREPORT-XML schema includes and extends the core VarioML schema:
datatypes xsd = "http://www.w3.org/2001/XMLSchema-datatypes"
here .
[back to
top ]
Examples of VREPORT-XML submissions
VREPORT XML
Example 1
VREPORT replaces individual with panel , which specifies a population of individuals.
<vreport
xmlns="http://gen2phen.org/varioml/2.0"
xmlns:xlink="http://www.w3.org/1999/xlink"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:noNamespaceSchemaLocation="vreports.xsd"
>
<source id="exdb">
<name>Example database</name>
<contact>
<name>Mr. Example</name>
</contact>
</source>
<panel id="0001">
<frequency type="carrier">
<category term="exist"/>
</frequency>
<population term="Finland" type="region"/>
<variant id="1002">
<gene accession="ACLY"/>
<ref_seq accession="n123445"/>
<name scheme="HGVS">c.12345G>A</name>
</variant>
</panel>
</vreport>
[back to top ]
VREPORT-XML Example: VarioML for general Variant Reports
<vreport
[back to
top ]
VREPORT NEMDB Example
This more complex example shows how VREPORT extends VarioML for use with National and Ethnic Mutation Databases (NEMDBs).
<?xml version='1.0' encoding='UTF-8'?>
<vreport
xmlns="http://gen2phen.org/varioml/2.0"
xmlns:xlink="http://www.w3.org/1999/xlink"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:noNamespaceSchemaLocation="vreports.xsd"
>
<source >
<name>FINDbase-PGx</name>
<contact>
<name>George Patrinos</name>
</contact>
</source>
<panel>
<frequency samples="50" type="allele">
<freq>0.06</freq>
</frequency>
<population type="group" term="Yorubans"/>
<population type="region" term="Nigeria"/>
<variant>
<gene accession="CYP3A5"/>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
<db_xref source="dbsnp" accession="rs776746"/>
<comment term="name"><text>CYP3A5*3</text></comment>
</variant>
</panel>
<!--
<panel>
<frequency samples="224" type="allele">
<freq>0.763</freq>
</frequency>
<population type="group" term="El Salvadorans"/>
<population type="ethnic" term="Mestizo">
<comment><text>Amerindian and European descent</text></comment>
</population>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.1289T>G</name>
</variant>
</panel>
<!--
Note that variant can also contain a variant specific frequency .
<panel>
<population type="group" term="Yorubans"/>
<population type="region" term="Nigeria"/>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
<frequency samples="50" type="allele">
<freq>0.06</freq>
</frequency>
</variant>
</panel>
<panel>
<frequency samples="66" type="allele">
<freq>0.11</freq>
</frequency>
<population type="group" term="Baka Pygmies"/>
<population type="region" term="Central African Republic"/>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
<panel>
<frequency samples="640" type="allele">
<freq>0.145</freq>
</frequency>
<population type="group" term="South Africans"/>
<population type="region" term="Transkei area of Eastern and Western Cape"/>
<population type="language_family" term="Xhosa"/>
<population type="race" term="Black"/>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
<panel xmlns="http://gen2phen.org/varioml/2.0" >
<frequency samples="196" type="allele">
<freq>0.145</freq>
</frequency>
<population type="group" term="Malays"/>
<population type="ethnic" term="Singapore Malays mainly from the Malay Archipelago"/>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
<panel>
<frequency samples="222" type="allele">
<freq>0.64</freq>
</frequency>
<population type="group" term="Brazilians"/>
<population type="ethnic" term="'Pardo'">
<comment><text>brown/intermediate</text></comment>
</population>
<population type="region" term="Rio de Janeiro"></population>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
<panel >
<frequency samples="224" type="allele">
<freq>0.763</freq>
</frequency>
<population type="group" term="El Salvadorans"/>
<population type="ethnic" term="Mestizo">
<comment><text>Amerindian and European descent</text></comment>
</population>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
<panel>
<frequency samples="176" type="allele">
<freq>0.926</freq>
</frequency>
<population type="group" term="Yakuts"/>
<population type="region" term="Village of Cheriktei in the Sakha (Yakutia) Republic">
</population>
<variant>
<ref_seq accession="NM_000777"/>
<name>g.6986A>G</name>
</variant>
</panel>
</vreport>
[back to top ]
VarioML classes and
properties cross-reference VarioML introduces the following classes and properties:
See the
VarioML
schema for more
details.
Schema Document
Properties | Global Declarations
| Global Definitions
Target Namespace
http://gen2phen.org/varioml/1.0
Element and Attribute Namespaces
Global element and attribute declarations belong to this schema's target namespace.
By default, local element declarations belong to this schema's target namespace.
By default, local attribute declarations have no namespace.
Declared Namespaces
Prefix
Namespace
xml
http://www.w3.org/XML/1998/namespace
vml
http://gen2phen.org/varioml/1.0
xs
http://www.w3.org/2001/XMLSchema
Schema Component Representation
<
xs:schema elementFormDefault ="
qualified "
targetNamespace ="
http://gen2phen.org/varioml/1.0 ">
...
</
xs:schema >
Name
acknowledgement
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Acknowledgement.
Schema Component Representation
<
xs:element name ="
acknowledgement ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Schema Component Representation
<
xs:element name ="
address "
type ="
xs :string"/>
Name
aliases
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Aliases or alternative legacy definitions (names) of the variant.
Element has list of sequence variants (recursive data structure) which can be seen as aliases of the main variant. Typically the variants are annotated against different reference sequences, and/or their name is based on a different naming scheme.
Example:
<variant>
<ref_seq source="genbank" accession="NG_001337.2"/>
<name scheme="HGVS"> g.22048C>G</name>
<aliases>
<variant>
<ref_seq accession="NG_002335.2"/>
<name> g.1400G>C</name>
</variant>
<variant>
<ref_seq accession="NG_007072.2"/>
<name> g.14034G>C</name>
</variant>
<variant>
<ref_seq accession="NG_007088.1"/>
<name> g.28259G>C</name>
</variant>
</aliases>
<db_xref source="dbsnp" accession="rs13"/>
</variant>
XML Instance Representation
<
vml :aliases>
Start Group: vml :variant [1..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :aliases>
Schema Component Representation
<
xs:element name ="
aliases ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Element: call
Schema Component Representation
<
xs:element name ="
call "
type ="
xs :string"/>
This element can be used wherever the following element is referenced:
XML Instance Representation
<
vml :category
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </
vml :category>
Schema Component Representation
<
xs:element name ="
category "
substitutionGroup ="
vml:frequency_value ">
<
xs:complexType >
</
xs:complexType >
</
xs:element >
Element: chr
Schema Component Representation
<
xs:element name ="
chr "
type ="
xs :string"/>
Element:
Name
comment
Type
vml :VmlComment
Nillable
no
Abstract
no
Documentation
Comments are of type Evidence_ontology. The term 'attribute' is optional. See under description of Evidence_ontology_term data type.
Comments can contain other comments.
Examples:
<comment>
<text> simple comment</text>
</comment>
<comment source="example_ontology" term="example term">
<text> my defined comment</text>
</comment>
<comment>
<text> whatever</text>
<comment>
<text> my structured comment</text>
</comment>
</comment>
<comment>
<text> whatever</text>
<comment>
<text> my complex structured comment</text>
<evidence_code term="something"/>
<db_xref accession="xyz"/>
</comment>
</comment>
<phenotype term="Autoimmune polyglandular syndrome type 1">
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
Schema Component Representation
Name
consent
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
NOT USED
Database cross-reference for a consent information (not used currently)
XML Instance Representation
<
vml :consent
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :consent>
Schema Component Representation
<
xs:element name ="
consent ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
consequence
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Consequences of mutation on sequence or structural levels.
lsdb-controlled-vocabulary-terms
Complex frameshift: Frameshift involving insertions and deletions
Exon deletion: Deletion encompassing a whole exon or exons, frameshift status unknown
Exon duplication: Duplication of one of more exons, frameshift status unknown
Frameshift: Deletion or insertion causing reading frame shift
In-frame deletion: Deletion of a whole codon or codons. Can include deletion of one or more exons
In-frame duplication: A duplication that does not change the reading frame. Can include one or more exons
In-frame insertion: An insertion of a whole codon or codons. Can include one or more exons
Intronic variant: A variant in an intron which has not been shown to affect splicing
Missense: Substitution resulting in a change to a different amino acid
Nonsense: Substitution resulting in a change to a stop codon
Out of frame deletion: Deletion of part of a codon or number of codons resulting in a frameshift. Can include one or more exons
Out of frame duplication: A duplication that changes the reading frame. Can include one or more exons
Out of frame insertion: An insertion of part of a codon or number of codons resulting in a frameshift. Can include one or more exons
Silent: A nucleotide change that does not change the amino acid
Splice site variant: A mutation that affects splicing
Example:
<consequence sourcels="SO" accession="1000071" term="mutation affecting splicing"/>
XML Instance Representation
<
vml :consequence
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :consequence>
Schema Component Representation
<
xs:element name ="
consequence ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Schema Component Representation
<
xs:element name ="
contact ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
This element can be used wherever the following element is referenced:
Schema Component Representation
<
xs:element name ="
counts "
type ="
xs :int"
substitutionGroup ="
vml:frequency_value "/>
Name
created
Type
xs :dateTime
Nillable
no
Abstract
no
Documentation
Submission/creation time stamp
XML Instance Representation
<
vml :created>
xs :dateTime </
vml :created>
Schema Component Representation
<
xs:element name ="
created "
type ="
xs :dateTime"/>
Name
cultivar
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Cultivar
XML Instance Representation
<
vml :cultivar
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :cultivar>
Schema Component Representation
<
xs:element name ="
cultivar ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
db_xref
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Abstract data type for database cross references.
Can include other database cross-references and annotations.
Examples:
<db_xref source="pubmed" accession="16965330"/>
XML Instance Representation
<
vml :db_xref
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :db_xref>
Schema Component Representation
<
xs:element name ="
db_xref ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: dob
Schema Component Representation
<
xs:element name ="
email "
type ="
xs :string"/>
XML Instance Representation
<
vml :embargo_end_date
is_undefined="xs :tokenvalue comes from list: {'true'})[0..1] " >
xs :date </
vml :embargo_end_date>
Element: end
Schema Component Representation
<
xs:element name ="
end "
type ="
xs :integer"/>
Name
evidence_code
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Evidence code is an ontology term (i.e. has optional source and accession attributes), which provides information related to the validity or the quality of observation.
Examples:
<evidence_code term="inferred"/>
<evidence_code term="confirmed" source="vario"/>
dbSNP validation should be used for mutations
Not validated (accession=0), Multiple reporting (1), With frequency
(2),Both frequency (3), Submitter validation (4)
XML Instance Representation
<
vml :evidence_code
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :evidence_code>
Schema Component Representation
<
xs:element name ="
evidence_code ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: exon
Name
exon
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Exon has optional attributes source ,accession and
transcript_ref which can be used for crossreference purposes. Exon number is given in text node
Example:
<exon> 01</exon>
XML Instance Representation
<
vml :exon
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " transcript_ref="anySimpleType [0..1] " />
Schema Component Representation
<
xs:element name ="
exon ">
<
xs:complexType mixed ="
true ">
</
xs:complexType >
</
xs:element >
Element: fax
Schema Component Representation
<
xs:element name ="
fax "
type ="
xs :string"/>
Element: freq
This element can be used wherever the following element is referenced:
Schema Component Representation
<
xs:element name ="
freq "
type ="
xs :decimal"
substitutionGroup ="
vml:frequency_value "/>
Name
frequency
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Actual frequency is stored in a subelement depending on value type. This type can be: decimal number, number of cases, or categorized value.
Frequency has the optional attribute 'samples', which can hold information on sample size. Typically this is the number of chromosomes. The calculation method type should be cross-referenced using protocol_id.
Frequency type can be either "allele" or "carrier", with optional subelements:
population
Evidence_ontology_term-type
evidence code
protocol id
comment
Examples:
<frequency samples="1000">
<population type="nationality" term="Finnish"/>
<freq> 0.2</freq>
</frequency>
<frequency> <freq> 0.1</freq></frequency>
<frequency samples="1000">
<cases> 11</cases>
</frequency>
<frequency>
<population type="region" term="Sweden"/>
<category term="exist"/>
</frequency>
<frequency samples="1111">
<population type="ethnic" term="Caucasian"/>
<freq> 0.12</freq>
<evidence_code source="sampling_ontology" accession="abcd" term="random"/>
</frequency>
XML Instance Representation
<
vml :frequency
samples="xs :int[0..1] " type="xs :tokenvalue comes from list: {'allele'|'carrier'|'genotype'})[0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :frequency>
Schema Component Representation
<
xs:element name ="
frequency ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
The following elements can be used wherever this element is referenced:
XML Instance Representation
<
vml :frequency_value> ... </
vml :frequency_value>
Name
gender
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Gender of individual. Gender code is stored in the attribute
'code'
. Possible values are iso5218 codes: 0,1,2 and 9 ('unknown','male','female' and 'not applicable').
Description of gender (code: 9) can be stored into optional
'description'
-sub-element, which is of type
Evidence_ontology_term
.
Example:
<gender code="9">
<description term="XX-male"/>
</gender>
Schema Component Representation
<
xs:element name ="
gender ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: gene
Name
gene
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Database cross reference where 'source' is the name of the database or system (e.g. HUGO) and 'accession' is the gene name (e.g.AGA).
HGNC names (symbols) should be used to name genes.
Can include other database cross-references and annotations.
Example source:
Example accession:
Examples:
<gene source="HGNC" accession="BRACA2"/>
<gene source="HGNC" accession="BRACA2">
<db_xref accession="600185" uri="http://www.ncbi.nlm.nih.gov/omim/600185" source="OMIM"/>
<db_xref accession="BRACA2" source="HGNC" uri="http://www.genenames.org/data/hgnc_data.php?hgnc_id=1101"/>
</gene>
XML Instance Representation
<
vml :gene
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :gene>
Schema Component Representation
<
xs:element name ="
gene ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
genetic_origin
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Genetic origin of variation. Has following information:
term - ontology term for origin of variant (denovo, sporadic, inherited)
source - parental origin of variation (paternal, maternal)
evidence code - evidence code for the observation
list of individuals
Genetic origin and source are evidence ontology terms.
Controlled vocabulary terms for genetic origin:
lsdb-controlled-vocabulary-terms
inherited
sporadic
de novo
Example:
<genetic_origin term="inherited">
<source term="paternal"/>
<evidence_code term="inferred"/>
</genetic_origin>
Schema Component Representation
<
xs:element name ="
genetic_origin ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
genotype
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Genotype
Schema Component Representation
<
xs:element name ="
genotype ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
XML Instance Representation
<
vml :grant_number
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</
vml :grant_number>
Name
group_type
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Group type
XML Instance Representation
<
vml :group_type
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :group_type>
Schema Component Representation
<
xs:element name ="
group_type ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
individual
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Individual or patient
Example:
<individual>
<gender code="2"/>
<phenotype term="Osteogenesis Imperfecta">
</phenotype>
<variant type="cDNA">
<gene source="HGNC" accession="COL1A1"/>
<ref_seq accession="NG_007400.1"/>
<name scheme="HGVS"> c.579delT</name>
<genetic_origin term="paternal">
<evidence_code term="inferred"/>
</genetic_origin>
<sharing_policy type="openAccess">
<embargo_end_date> 2002-12-12</embargo_end_date>
<use_permission accession="CC0" uri="http://creativecommons.org/publicdomain/zero/1.0/" term="Creative commons"/>
</sharing_policy>
<comment>
<text> Variant inherited from affected father</text>
</comment>
</variant>
</individual>
XML Instance Representation
<
vml :individual
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :variant [0..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :individual>
Schema Component Representation
<
xs:element name ="
individual ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
inheritance_pattern
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Inheritance pattern of the phenotype. Possible values for the term:
familial
familial, consanguineous parents
familial, autosomal dominant
familial, autosomal recessive
familial, Xlinked
sporadic
sporadic, consanguineous parents
sporadic, consanguineous parents (1st degree)
sporadic, consanguineous parents (2nd degree)
sporadic, consanguineous parents (3rd degree)
sporadic, nonconsanguineous parents
sporadic, consanguinity parents?
sporadic? (parents not tested)
See:
lsdb-controlled-vocabulary-terms
XML Instance Representation
<
vml :inheritance_pattern
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :inheritance_pattern>
Schema Component Representation
<
xs:element name ="
inheritance_pattern ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
location
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Location of variant on a reference sequence/genomic build
Example:
<location>
<ref_seq source="embl" accession="ABC1023345"/>
<start> 111111</start>
<end> 111112</end>
</location>
<location>
<ref_seq accession="build36"/>
<chr> 21</chr>
<start> 111111</start>
<end> 111112</end>
</location>
Schema Component Representation
<
xs:element name ="
location ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Element: lsdb
Name
lsdb
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Single data submission batch element for individual data. The batch contains:
id,uri - optional submission attributes
created - optional timestamp when data submission batch was created
source - information on source(s) of submission. Submitting source
list of individuals
Example:
<lsdb xsi:noNamespaceSchemaLocation="lsdb.xsd">
<created> 2000-01-12T12:13:14Z</created>
<source id="AIREbase" version="1.2" uri="http://www.uta.fi/imt/bioinfo/AIREbase/">
<name> DatabaseXYZ</name>
<url> http://www.uta.fi/imt/bioinfo/AIREbase/</url>
<contact>
<name> Matti Meik?l?inen</name>
<address> Institute for Molecular Medicine</address>
</contact>
<acknowledgement>
<name> European Union</name>
<grant_number accession="23230001"/>
</acknowledgement>
</source>
<individual id="xyz">
<gender code="2"/>
<variant>
<ref_seq accession="xyz"/>
<name scheme="HGVS"> c.1343G>T</name>
</variant>
</individual>
<individual id="abce">
<gender code="2"/>
<variant>
<ref_seq accession="xyz"/>
<name scheme="HGVS"> c.1343G>C</name>
</variant>
</individual>
</lsdb>
Schema Component Representation
<
xs:element name ="
lsdb ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
modification_date
Type
vml :VmlModificationDate
Nillable
no
Abstract
no
Documentation
Date when submitted information was modified.
Example:
<modification_date> 2001</modification_date>
<modification_date> 2001-09</modification_date>
<modification_date> 2001-09-10</modification_date>
Name
observations
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Abstract type. See subtypes for the documentation. Observation which has evidence code and observation protocol (dbxref).
Example:
<genetic_origin term="paternal">
<evidence_code term="inferred"/>
</genetic_origin>
<phenotype term="Osteogenesis Imperfecta">
<evidence_code term="assessed by clinician"/>
<protocol_id source="protocoldb_xyz" accession="XYZ0001"/>
</phenotype>
XML Instance Representation
<
vml :observations>
Start Choice [0..1] End Choice Start Group: vml :variant [0..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :observations>
Schema Component Representation
<
xs:element name ="
observations ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Name
organism
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Organism
XML Instance Representation
<
vml :organism
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :organism>
Schema Component Representation
<
xs:element name ="
organism ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
original_id
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Identifier used by the submitter.
XML Instance Representation
<
vml :original_id
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :original_id>
Schema Component Representation
<
xs:element name ="
original_id ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
panel
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Panel. Unspecified group of individuals. Has following information besides properties inherited from AbstractObservation target:
frequency - frequency of given variant or variant set
individuals - list of individuals beloning to the panel
Example:
Frequency for existence of two alleles.
<panel>
<frequency samples="224" type="allele">
<freq> 0.763</freq>
</frequency>
<population type="group" term="El Salvadorans"/>
<population type="ethnic" term="Mestizo">
<comment> <text> Amerindian and European descent</text></comment>
</population>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.6986A>G</name>
</variant>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.1289T>G</name>
</variant>
</panel>
Note also that also variant specific frequency can be given as usual:
<panel>
<population type="group" term="Yorubans"/>
<population type="region" term="Nigeria"/>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.6986A>G</name>
<frequency samples="50" type="allele">
<freq> 0.06</freq>
</frequency>
</variant>
</panel>
Usage of the format must be agreed on separately.
XML Instance Representation
<
vml :panel
size="xs :integer[0..1] " type="xs :tokenvalue comes from list: {'family'|'extended family'|'population'})[0..1] " id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :variant [0..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :panel>
Schema Component Representation
<
xs:element name ="
panel ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
phenotype
Type
vml :VmlPhenotype
Nillable
no
Abstract
no
Documentation
Phenotype of individual. Phenotype is evidence ontology term (see evidence_ontology)
Example:
<phenotype term="Autoimmune polyglandular syndrome type 1"/>
<phenotype term="Autoimmune polyglandular syndrome type 1" accession="240300" source="OMIM">
<evidence_code term="confirmed" source="vario"/>
<protocol_id accession="observation_method_xyz" source="protocol_database"/>
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
<
vml :phenotype
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :phenotype>
Schema Component Representation
<
xs:element name ="
phone "
type ="
xs :string"/>
Name
population
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Information related to population group the individual belongs to.
Population element has the "type" attribute which indicates the selection criteria of the population, using the values:
ethnic
region
race
other
unknown
The attribute is informative: the actual type of population should be defined in the source ontology (if it is used).
Example:
<population type="ethnic" term="Caucasian"/>
<population type="ethnic" term="Caucasian">
<evidence_code source="example_ontology_term" accession="abcd" term="ethnicity of mother"/>
</population>
<population type="ethnic" term="Asian">
<evidence_code source="example_ontology_term" accession="abcd" term="ethnicity of father"/>
</population>
<population type="region" term="Ahvenanmaa" source="ISO 3166" accession="ISO 3166-2:AX">
<evidence_code term="place of birth"/>
</population>
XML Instance Representation
<
vml :population
type="xs :tokenvalue comes from list: {'region'|'nationality'|'ethnic'|'race'|'organism'|'group'|'primary_language'|'language_family'|'other'|'unknown'})[1] source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :population>
Schema Component Representation
<
xs:element name ="
population ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
protocol_id
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Cross-reference to a protocol database.
XML Instance Representation
<
vml :protocol_id
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :protocol_id>
Schema Component Representation
<
xs:element name ="
protocol_id ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
ref_seq
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Database cross reference for reference sequence.
Can include other database cross-references and annotations.
Example:
<ref_seq source="genbank" accession="NG_007088.1"/>
XML Instance Representation
<
vml :ref_seq
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :ref_seq>
Schema Component Representation
<
xs:element name ="
ref_seq ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
XML Instance Representation
<
vml :reference>
xs :string </
vml :reference>
Schema Component Representation
<
xs:element name ="
reference "
type ="
xs :string"/>
Name
relationship
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
relationships .. NOT NEEDED
Relationship
XML Instance Representation
<
vml :relationship
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :relationship>
Schema Component Representation
<
xs:element name ="
relationship ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
restriction_site
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Restriction enzyme site.
XML Instance Representation
<
vml :restriction_site
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :restriction_site>
Schema Component Representation
<
xs:element name ="
restriction_site ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: role
Name
role
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Role
XML Instance Representation
<
vml :role
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :role>
Schema Component Representation
<
xs:element name ="
role ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
sample
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Sample
XML Instance Representation
<
vml :sample
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :sample>
Schema Component Representation
<
xs:element name ="
sample ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
score
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Score
XML Instance Representation
<
vml :score
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :score>
Schema Component Representation
<
xs:element name ="
score ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
seq_changes
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Related sequence changes on overlapping sequence levels and features.
Element has recursive list of sequence variants of type RNA or AA. The Sub-variant element differ from the main variant element in a following way:
type has only values RNA and AA
ref_seq is optional
gene is missing
XML Instance Representation
<
vml :seq_changes>
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'cDNA'|'RNA'|'AA'})[0..1] " >
[1..*] <
vml :name
scheme="anySimpleType [0..1] " />
[1] Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :seq_changes>
Schema Component Representation
<
xs:element name ="
seq_changes ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Name
sequence
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Reference and variant sequence strings. Accession number of reference sequence is given in ref_seq element (see the variant)
Example:
<sequence>
<reference> atttgatcgttc</reference>
<variant> atttgGtcgttc</variant>
</sequence>
Schema Component Representation
<
xs:element name ="
sequence ">
<
xs:complexType >
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
</
xs:element >
Name
sequence_region
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Name of the sequence region (not used)
XML Instance Representation
<
vml :sequence_region
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :sequence_region>
Schema Component Representation
<
xs:element name ="
sequence_region ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
sharing_policy
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Data sharing policy. Following access policy types are used:
closedAccess,embargoedAccess,restrictedAccess and openAccess
Adopted from OpenAIRE : specification
Example:
<sharing_policy type="openAccess">
<embargo_end_date> 2002-12-12</embargo_end_date>
<use_permission accession="CC0" uri="http://creativecommons.org/publicdomain/zero/1.0/" term="Creative commons"/>
</sharing_policy>
XML Instance Representation
<
vml :sharing_policy
type="xs :tokenvalue comes from list: {'closedAccess'|'embargoedAccess'|'restrictedAccess'|'openAccess'})[1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :sharing_policy>
Schema Component Representation
<
xs:element name ="
sharing_policy ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Schema Component Representation
<
xs:element name ="
start "
type ="
xs :integer"/>
Name
strain
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Strain
XML Instance Representation
<
vml :strain
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :strain>
Schema Component Representation
<
xs:element name ="
strain ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
submitter_id
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Randomized identifier of individual as used by submitter.
XML Instance Representation
<
vml :submitter_id
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :submitter_id>
Schema Component Representation
<
xs:element name ="
submitter_id ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: text
Name
text
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Text
XML Instance Representation
<
vml :text
content_type="anySimpleType [0..1] " lang="anySimpleType [0..1] " encoding="anySimpleType [0..1] " />
Schema Component Representation
<
xs:element name ="
text ">
<
xs:complexType mixed ="
true ">
</
xs:complexType >
</
xs:element >
Name
tissue
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Tissue
XML Instance Representation
<
vml :tissue
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :tissue>
Schema Component Representation
<
xs:element name ="
tissue ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
tissue_distribution
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Tissue distribution
Controlled vocabulary terms for tissue disttribution:
lsdb-controlled-vocabulary-terms
constitutional
mosaic
mosaic in germline
Example:
<tissue_distribution term="mosaic"/>
XML Instance Representation
<
vml :tissue_distribution
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :tissue_distribution>
Schema Component Representation
<
xs:element name ="
tissue_distribution ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Element: url
Schema Component Representation
<
xs:element name ="
url "
type ="
xs :string"/>
Name
use_permission
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Licensing information.
Example:
<use_permission accession="CC0" uri="http://creativecommons.org/publicdomain/zero/1.0/" term="Creative commons"/>
XML Instance Representation
<
vml :use_permission
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :use_permission>
Schema Component Representation
<
xs:element name ="
use_permission ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Schema Component Representation
<
xs:element name ="
value "
type ="
xs :float"/>
XML Instance Representation
<
vml :variant_class
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant_class>
Schema Component Representation
<
xs:element name ="
variant_class ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
variant_detection
Type
vml :VmlVariantDetection
Nillable
no
Abstract
no
Documentation
Techniques used to detect the variant.
Variant detection method. Element has attributes:
sequence template, which can be either DNA, RNA, cDNA or AA
detection technique
Optional element protocol_id (of type DbXRef) has reference to actual detection protocol.
Controlled vocabulary terms for detection techniques:
lsdb-controlled-vocabulary-terms
ARMS
CF20: CF Common Mutation Test
CF29: Analysis of 29 mutations using the Elucigene CF29 kit
CSCE: Conformation sensitive capillary electrophoresis
DGGE: Denaturing gradient gel electrophoresis
dHPLC: Denaturing high performance liquid chromatography
Heteroduplex analysis
Loss of heterozygosity analysis
Meta-PCR
MLPA: Multiplex ligation-dependent probe amplification
MS-PCR: Mutagenically separated PCR
Multiplex PCR
Not Known: The information has not been recorded or provided
Not Specified: Test information cannot be determined
PCR-PAGE
PTT: Protein Trucation Test
RNA: RNA work performed
Sequencing
SNPlex: The SNPlex™ Genotyping System from ABI
SSCP
SSCP/Heteroduplex
Example:
<variant_detection template="DNA" technique="DHPLC"/>
XML Instance Representation
<
vml :variant_detection
template="xs :tokenvalue comes from list: {'DNA'|'RNA'|'cDNA'|'AA'})[1] " technique="anySimpleType [1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant_detection>
Name
variant_group
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
Variant group
XML Instance Representation
<
vml :variant_group
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " orientation="xs :tokenvalue comes from list: {'cis'|'trans'|'unknown'})[0..1] " >
Start Group: vml :variant [1..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant_group>
Schema Component Representation
<
xs:element name ="
variant_group ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
variant_type
Type
Locally-defined complex type
Nillable
no
Abstract
no
Documentation
See types of sequence alteration in Sequence Ontology
XML Instance Representation
<
vml :variant_type
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant_type>
Schema Component Representation
<
xs:element name ="
variant_type ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
Name
VmlAbstractObservationTarget
Documentation
Abstract observation target. Contains information which is common to all observation targets like individuals and panels.
id,uri - optional unqiue identifiers
original_id - optional source identifier as used in source system
organism
phenotypes -list of phenotypes
populations - list of population groups target belongs to
variants -list of variants targes has
source - original source of the information
Schema Component Representation
<
xs:attributeGroup name ="
VmlAbstractObservationTarget ">
</
xs:attributeGroup >
Name
VmlAnything
Documentation
Anything.
Schema Component Representation
<
xs:attributeGroup name ="
VmlAnything ">
<xs:anyAttribute processContents ="skip "/>
</
xs:attributeGroup >
Name
VmlEvidenceOntologyTerm
Documentation
Abstract datatype (superclass) for controlled vocabulary terms related to observations. All Evidence_ontology_term elements have:
optional protocol and database crossreferences - list of protocol_id and db_xref elements
evidence codes - list of evidence_code-elements
In addition of properties inherited from ontology_class (like db_xref and comment)
<genetic_origin term="inherited">
<source term="paternal"/>
<evidence_code term="inferred"/>
</genetic_origin>
<phenotype term="Osteogenesis Imperfecta">
<protocol_id accession="observation_methodXYZ" source="protocol_database"/>
</phenotype>
<phenotype term="Autoimmune polyglandular syndrome type 1">
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
Schema Component Representation
<
xs:attributeGroup name ="
VmlEvidenceOntologyTerm ">
</
xs:attributeGroup >
Attribute Group: VmlExon
Name
VmlExon
Documentation
Exon has optional attributes source ,accession and
transcript_ref which can be used for crossreference purposes. Exon number is given in text node
Example:
<exon> 01</exon>
Schema Component Representation
<
xs:attributeGroup name ="
VmlExon ">
<xs:attribute name ="source "/>
<xs:attribute name ="accession "/>
<xs:attribute name ="transcript_ref "/>
</
xs:attributeGroup >
Name
VmlForeignAttributes
Documentation
Define foreign nodes for extensions.
Schema Component Representation
<
xs:attributeGroup name ="
VmlForeignAttributes ">
<xs:anyAttribute namespace ="##other " processContents ="skip "/>
</
xs:attributeGroup >
Name
VmlForeignNodes
Documentation
Define foreign nodes for extensions.
Schema Component Representation
<
xs:attributeGroup name ="
VmlForeignNodes ">
</
xs:attributeGroup >
Name
VmlIdentifiable
Documentation
Abstract class for identifiables
id - unqiue id
uri - uri of the identifiable
Schema Component Representation
<
xs:attributeGroup name ="
VmlIdentifiable ">
<
xs:attribute name ="
id "
type ="
vml :VmlId "/>
</
xs:attributeGroup >
Attribute Group: VmlName
Name
VmlName
Documentation
Name of variant
Element has following optional attributes:
scheme
which is naming scheme which should typically be HGVS.
Example:
<name type="DNA" scheme="HGVS">
c.755G\>A
</name>
Schema Component Representation
<
xs:attributeGroup name ="
VmlName ">
<xs:attribute name ="scheme "/>
</
xs:attributeGroup >
Name
VmlOntology
Documentation
Ontology class. Supertype for ontology terms (most of the VarioML elements are ontology or evidence ontology terms)
source - name (abbreviation) of system where term is defined e.g. SO (sequence ontology)
accession - accession number of term
term - ontology term
uri - uri of the term
dbx_ref - database cross-references
comment - comments
Example:
<phenotype term="Autoimmune polyglandular syndrome type 1" accession="240300" source="OMIM">
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
Schema Component Representation
<
xs:attributeGroup name ="
VmlOntology ">
<xs:attribute name ="source "/>
<xs:attribute name ="accession "/>
<xs:attribute name ="term " use ="required "/>
</
xs:attributeGroup >
Name
VmlOntologyTerm
Documentation
Abstract annotated ontology class. See supertype for more information.
Schema Component Representation
<
xs:attributeGroup name ="
VmlOntologyTerm ">
</
xs:attributeGroup >
Attribute Group: VmlText
Name
VmlText
Documentation
Text
Schema Component Representation
<
xs:attributeGroup name ="
VmlText ">
<xs:attribute name ="content_type "/>
<xs:attribute name ="lang "/>
<xs:attribute name ="encoding "/>
</
xs:attributeGroup >
Complex Type:
Super-types:
None
Sub-types:
None
Name
VmlComment
Abstract
no
Documentation
Comments are of type Evidence_ontology. The term 'attribute' is optional. See under description of Evidence_ontology_term data type.
Comments can contain other comments.
Examples:
<comment>
<text> simple comment</text>
</comment>
<comment source="example_ontology" term="example term">
<text> my defined comment</text>
</comment>
<comment>
<text> whatever</text>
<comment>
<text> my structured comment</text>
</comment>
</comment>
<comment>
<text> whatever</text>
<comment>
<text> my complex structured comment</text>
<evidence_code term="something"/>
<db_xref accession="xyz"/>
</comment>
</comment>
<phenotype term="Autoimmune polyglandular syndrome type 1">
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
Schema Component Representation
Super-types:
VmlDbXRef VmlConsent (by extension)
Sub-types:
None
Name
VmlConsent
Abstract
no
Documentation
NOT USED
Database cross-reference for a consent information (not used currently)
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlConsent ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlConsequence
Abstract
no
Documentation
Consequences of mutation on sequence or structural levels.
lsdb-controlled-vocabulary-terms
Complex frameshift: Frameshift involving insertions and deletions
Exon deletion: Deletion encompassing a whole exon or exons, frameshift status unknown
Exon duplication: Duplication of one of more exons, frameshift status unknown
Frameshift: Deletion or insertion causing reading frame shift
In-frame deletion: Deletion of a whole codon or codons. Can include deletion of one or more exons
In-frame duplication: A duplication that does not change the reading frame. Can include one or more exons
In-frame insertion: An insertion of a whole codon or codons. Can include one or more exons
Intronic variant: A variant in an intron which has not been shown to affect splicing
Missense: Substitution resulting in a change to a different amino acid
Nonsense: Substitution resulting in a change to a stop codon
Out of frame deletion: Deletion of part of a codon or number of codons resulting in a frameshift. Can include one or more exons
Out of frame duplication: A duplication that changes the reading frame. Can include one or more exons
Out of frame insertion: An insertion of part of a codon or number of codons resulting in a frameshift. Can include one or more exons
Silent: A nucleotide change that does not change the amino acid
Splice site variant: A mutation that affects splicing
Example:
<consequence sourcels="SO" accession="1000071" term="mutation affecting splicing"/>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlConsequence ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlConsequentVariant
Abstract
no
Documentation
Corresponding variant on RNA or AA levels. Has same content as variant element except gene element is missing and ref_seq is optional.
XML Instance Representation
<...
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'cDNA'|'RNA'|'AA'})[0..1] " >
<
vml :name
scheme="anySimpleType [0..1] " />
[1] Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity </...>
Schema Component Representation
<
xs:complexType name ="
VmlConsequentVariant ">
<
xs:sequence >
<
xs:element ref ="
vml :consequence "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :phenotype "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :location "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
type ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="cDNA "/>
<xs:enumeration value ="RNA "/>
<xs:enumeration value ="AA "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlContact
Abstract
no
Documentation
Name and address of contact person
XML Instance Representation
<...
role="xs :string[0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlContact ">
<
xs:sequence >
<
xs:element name ="
name "
type ="
xs :string"/>
<
xs:element ref ="
vml :url "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :phone "
minOccurs ="
0 "/>
<
xs:element ref ="
vml :fax "
minOccurs ="
0 "/>
<
xs:element ref ="
vml :email "
minOccurs ="
0 "/>
</
xs:sequence >
<
xs:attribute name ="
role "
type ="
xs :string"/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlCultivar ">
</
xs:complexType >
Super-types:
None
Sub-types:
Name
VmlDbXRef
Abstract
no
Documentation
Email address
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlDbXRef ">
<xs:attribute name ="source "/>
<xs:attribute name ="accession " use ="required "/>
<xs:attribute name ="name "/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlDescription
Abstract
no
Documentation
Description of gender. Used when geneder code i 9
Example:
<gender code="9">
<description term="XX-male"/>
</gender>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlDescription ">
</
xs:complexType >
Super-types:
xs :dateVmlEmbargo_end_date (by extension)
Sub-types:
None
Name
VmlEmbargo_end_date
Abstract
no
Documentation
Embargo end date
XML Instance Representation
<...
is_undefined="xs :tokenvalue comes from list: {'true'})[0..1] " >
xs :date </...>
Schema Component Representation
<
xs:complexType name ="
VmlEmbargo_end_date ">
<
xs:simpleContent >
<
xs:extension base ="
xs :date">
<
xs:attribute name ="
is_undefined ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="true "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:extension >
</
xs:simpleContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlEvidenceCode ">
<
xs:sequence >
<
xs:element ref ="
vml :score "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlFactor
Abstract
no
Documentation
Pathogenicity of variant.
Pathogenicity has 7 possible values:
Non-pathogenic
Not known
Pathogenic
Probably not pathogenic
Probably pathogenic
Causative
Probably causative
Unclassified
Note: implementation of fixed vocabulary for this element is still pending. See:
lsdb-controlled-vocabulary-terms
Causative are added as workaround for cases where mutation do not lead to a disease phenotype (e.g. eye color etc.)
Causative is for cases where variant is known to have impact on non disease phenotype.
Pathogenicity has new attributes: phenotype, scope (individual, family, population) and score
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlFactor ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlFrequency
Abstract
no
Documentation
Actual frequency is stored in a subelement depending on value type. This type can be: decimal number, number of cases, or categorized value.
Frequency has the optional attribute 'samples', which can hold information on sample size. Typically this is the number of chromosomes. The calculation method type should be cross-referenced using protocol_id.
Frequency type can be either "allele" or "carrier", with optional subelements:
population
Evidence_ontology_term-type
evidence code
protocol id
comment
Examples:
<frequency samples="1000">
<population type="nationality" term="Finnish"/>
<freq> 0.2</freq>
</frequency>
<frequency> <freq> 0.1</freq></frequency>
<frequency samples="1000">
<cases> 11</cases>
</frequency>
<frequency>
<population type="region" term="Sweden"/>
<category term="exist"/>
</frequency>
<frequency samples="1111">
<population type="ethnic" term="Caucasian"/>
<freq> 0.12</freq>
<evidence_code source="sampling_ontology" accession="abcd" term="random"/>
</frequency>
XML Instance Representation
<...
samples="xs :int[0..1] " type="xs :tokenvalue comes from list: {'allele'|'carrier'|'genotype'})[0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlFrequency ">
<
xs:sequence >
<
xs:element ref ="
vml :population "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
samples "
type ="
xs :int"/>
<
xs:attribute name ="
type ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="allele "/>
<xs:enumeration value ="carrier "/>
<xs:enumeration value ="genotype "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlGender
Abstract
no
Documentation
Gender of individual. Gender code is stored in the attribute
'code'
. Possible values are iso5218 codes: 0,1,2 and 9 ('unknown','male','female' and 'not applicable').
Description of gender (code: 9) can be stored into optional
'description'
-sub-element, which is of type
Evidence_ontology_term
.
Example:
<gender code="9">
<description term="XX-male"/>
</gender>
XML Instance Representation
<...
code="xs :tokenvalue comes from list: {'0'|'1'|'2'|'9'})[1] " >
Start Group: vml :description [0..1] <
vml :description
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :description>
End Group: vml :description </...>
Schema Component Representation
<
xs:complexType name ="
VmlGender ">
<
xs:sequence >
</
xs:sequence >
<
xs:attribute name ="
code "
use ="
required ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="0 "/>
<xs:enumeration value ="1 "/>
<xs:enumeration value ="2 "/>
<xs:enumeration value ="9 "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
VmlDbXRef VmlGene (by extension)
Sub-types:
None
Name
VmlGene
Abstract
no
Documentation
Database cross reference where 'source' is the name of the database or system (e.g. HUGO) and 'accession' is the gene name (e.g.AGA).
HGNC names (symbols) should be used to name genes.
Can include other database cross-references and annotations.
Example source:
Example accession:
Examples:
<gene source="HGNC" accession="BRACA2"/>
<gene source="HGNC" accession="BRACA2">
<db_xref accession="600185" uri="http://www.ncbi.nlm.nih.gov/omim/600185" source="OMIM"/>
<db_xref accession="BRACA2" source="HGNC" uri="http://www.genenames.org/data/hgnc_data.php?hgnc_id=1101"/>
</gene>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlGene ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlGeneticSource
Abstract
no
Documentation
Genetic origin of variation. Has following information:
term - ontology term for origin of variant (denovo, sporadic, inherited)
source - parental origin of variation (paternal, maternal)
evidence code - evidence code for the observation
list of individuals
Genetic origin and source are evidence ontology terms.
Controlled vocabulary terms for genetic origin:
lsdb-controlled-vocabulary-terms
inherited
sporadic
de novo
Example:
<genetic_origin term="inherited">
<source term="paternal"/>
<evidence_code term="inferred"/>
</genetic_origin>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlGeneticSource ">
<
xs:sequence >
<
xs:element ref ="
vml :population "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlGenetic_origin
Abstract
no
Documentation
Genetic origin of variation. Has following information:
term - ontology term for origin of variant (denovo, sporadic, inherited)
source - parental origin of variation (paternal, maternal)
evidence code - evidence code for the observation
list of individuals
Genetic origin and source are evidence ontology terms.
Controlled vocabulary terms for genetic origin:
lsdb-controlled-vocabulary-terms
inherited
sporadic
de novo
Example:
<genetic_origin term="inherited">
<source term="paternal"/>
<evidence_code term="inferred"/>
</genetic_origin>
Schema Component Representation
<
xs:complexType name ="
VmlGenetic_origin ">
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlGroupType ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlIndividual
Abstract
no
Documentation
Individual or patient
Example:
<individual>
<gender code="2"/>
<phenotype term="Osteogenesis Imperfecta">
</phenotype>
<variant type="cDNA">
<gene source="HGNC" accession="COL1A1"/>
<ref_seq accession="NG_007400.1"/>
<name scheme="HGVS"> c.579delT</name>
<genetic_origin term="paternal">
<evidence_code term="inferred"/>
</genetic_origin>
<sharing_policy type="openAccess">
<embargo_end_date> 2002-12-12</embargo_end_date>
<use_permission accession="CC0" uri="http://creativecommons.org/publicdomain/zero/1.0/" term="Creative commons"/>
</sharing_policy>
<comment>
<text> Variant inherited from affected father</text>
</comment>
</variant>
</individual>
XML Instance Representation
<...
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :variant [0..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :source [0..1] End Group: vml :source </...>
Schema Component Representation
<
xs:complexType name ="
VmlIndividual ">
<
xs:sequence >
<
xs:element ref ="
vml :dob "
minOccurs ="
0 "/>
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlInheritancePattern ">
</
xs:complexType >
Super-types:
VmlSubmission VmlLsdb (by extension)
Sub-types:
None
Name
VmlLsdb
Abstract
no
Documentation
Single data submission batch element for individual data. The batch contains:
id,uri - optional submission attributes
created - optional timestamp when data submission batch was created
source - information on source(s) of submission. Submitting source
list of individuals
Example:
<lsdb xsi:noNamespaceSchemaLocation="lsdb.xsd">
<created> 2000-01-12T12:13:14Z</created>
<source id="AIREbase" version="1.2" uri="http://www.uta.fi/imt/bioinfo/AIREbase/">
<name> DatabaseXYZ</name>
<url> http://www.uta.fi/imt/bioinfo/AIREbase/</url>
<contact>
<name> Matti Meik?l?inen</name>
<address> Institute for Molecular Medicine</address>
</contact>
<acknowledgement>
<name> European Union</name>
<grant_number accession="23230001"/>
</acknowledgement>
</source>
<individual id="xyz">
<gender code="2"/>
<variant>
<ref_seq accession="xyz"/>
<name scheme="HGVS"> c.1343G>T</name>
</variant>
</individual>
<individual id="abce">
<gender code="2"/>
<variant>
<ref_seq accession="xyz"/>
<name scheme="HGVS"> c.1343G>C</name>
</variant>
</individual>
</lsdb>
Schema Component Representation
<
xs:complexType name ="
VmlLsdb ">
<
xs:complexContent >
<
xs:extension base ="
vml :VmlSubmission ">
<
xs:sequence >
</
xs:sequence >
</
xs:extension >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlOrganism ">
</
xs:complexType >
Super-types:
VmlDbXRef VmlOriginalId (by extension)
Sub-types:
None
Name
VmlOriginalId
Abstract
no
Documentation
Identifier used by the submitter.
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlOriginalId ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlPanel
Abstract
no
Documentation
Panel. Unspecified group of individuals. Has following information besides properties inherited from AbstractObservation target:
frequency - frequency of given variant or variant set
individuals - list of individuals beloning to the panel
Example:
Frequency for existence of two alleles.
<panel>
<frequency samples="224" type="allele">
<freq> 0.763</freq>
</frequency>
<population type="group" term="El Salvadorans"/>
<population type="ethnic" term="Mestizo">
<comment> <text> Amerindian and European descent</text></comment>
</population>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.6986A>G</name>
</variant>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.1289T>G</name>
</variant>
</panel>
Note also that also variant specific frequency can be given as usual:
<panel>
<population type="group" term="Yorubans"/>
<population type="region" term="Nigeria"/>
<variant>
<ref_seq accession="NM_000777"/>
<name> g.6986A>G</name>
<frequency samples="50" type="allele">
<freq> 0.06</freq>
</frequency>
</variant>
</panel>
Usage of the format must be agreed on separately.
XML Instance Representation
<...
size="xs :integer[0..1] " type="xs :tokenvalue comes from list: {'family'|'extended family'|'population'})[0..1] " id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " >
Start Group: vml :variant [0..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :source [0..1] End Group: vml :source </...>
Schema Component Representation
<
xs:complexType name ="
VmlPanel ">
<
xs:sequence >
<
xs:element ref ="
vml :individual "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
size "
type ="
xs :integer"/>
<
xs:attribute name ="
type ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="family "/>
<xs:enumeration value ="extended family "/>
<xs:enumeration value ="population "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlPathogenicity
Abstract
no
Documentation
Pathogenicity of variant.
Pathogenicity has 7 possible values:
Non-pathogenic
Not known
Pathogenic
Probably not pathogenic
Probably pathogenic
Causative
Probably causative
Unclassified
Note: implementation of fixed vocabulary for this element is still pending. See:
lsdb-controlled-vocabulary-terms
Causative are added as workaround for cases where mutation do not lead to a disease phenotype (e.g. eye color etc.)
Causative is for cases where variant is known to have impact on non disease phenotype.
Pathogenicity has new attributes: phenotype, scope (individual, family, population) and score
XML Instance Representation
<...
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlPathogenicity ">
<
xs:sequence >
<
xs:element ref ="
vml :phenotype "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:group ref ="
vml :factor "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
scope ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="individual "/>
<xs:enumeration value ="family "/>
<xs:enumeration value ="population "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
<
xs:attribute name ="
panel_ref "
type ="
vml :VmlId "/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlPhenotype
Abstract
no
Documentation
Phenotype of individual. Phenotype is evidence ontology term (see evidence_ontology)
Example:
<phenotype term="Autoimmune polyglandular syndrome type 1"/>
<phenotype term="Autoimmune polyglandular syndrome type 1" accession="240300" source="OMIM">
<evidence_code term="confirmed" source="vario"/>
<protocol_id accession="observation_method_xyz" source="protocol_database"/>
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </...>
Schema Component Representation
<
xs:complexType name ="
VmlPhenotype ">
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlPopulation
Abstract
no
Documentation
Information related to population group the individual belongs to.
Population element has the "type" attribute which indicates the selection criteria of the population, using the values:
ethnic
region
race
other
unknown
The attribute is informative: the actual type of population should be defined in the source ontology (if it is used).
Example:
<population type="ethnic" term="Caucasian"/>
<population type="ethnic" term="Caucasian">
<evidence_code source="example_ontology_term" accession="abcd" term="ethnicity of mother"/>
</population>
<population type="ethnic" term="Asian">
<evidence_code source="example_ontology_term" accession="abcd" term="ethnicity of father"/>
</population>
<population type="region" term="Ahvenanmaa" source="ISO 3166" accession="ISO 3166-2:AX">
<evidence_code term="place of birth"/>
</population>
XML Instance Representation
<...
type="xs :tokenvalue comes from list: {'region'|'nationality'|'ethnic'|'race'|'organism'|'group'|'primary_language'|'language_family'|'other'|'unknown'})[1] source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlPopulation ">
<
xs:attribute name ="
type "
use ="
required ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="region "/>
<xs:enumeration value ="nationality "/>
<xs:enumeration value ="ethnic "/>
<xs:enumeration value ="race "/>
<xs:enumeration value ="organism "/>
<xs:enumeration value ="group "/>
<xs:enumeration value ="primary_language "/>
<xs:enumeration value ="language_family "/>
<xs:enumeration value ="other "/>
<xs:enumeration value ="unknown "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
VmlDbXRef VmlProtocolId (by extension)
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlProtocolId ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
VmlDbXRef VmlRefSeq (by extension)
Sub-types:
None
Name
VmlRefSeq
Abstract
no
Documentation
Database cross reference for reference sequence.
Can include other database cross-references and annotations.
Example:
<ref_seq source="genbank" accession="NG_007088.1"/>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlRefSeq ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlRelationship ">
<
xs:sequence >
<
xs:element ref ="
vml :individual "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :panel "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlRestrictionSite
Abstract
no
Documentation
Restriction enzyme site.
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlRestrictionSite ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlRoleType
Abstract
no
Documentation
Subject Type
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlRoleType ">
</
xs:complexType >
Super-types:
VmlDbXRef VmlSample (by extension)
Sub-types:
None
Name
VmlSample
Abstract
no
Documentation
Sample
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlSample ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlScore ">
<
xs:sequence >
</
xs:sequence >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlSequenceRegion
Abstract
no
Documentation
Name of the sequence region (not used)
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlSequenceRegion ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlSharingPolicy
Abstract
no
Documentation
Data sharing policy. Following access policy types are used:
closedAccess,embargoedAccess,restrictedAccess and openAccess
Adopted from OpenAIRE : specification
Example:
<sharing_policy type="openAccess">
<embargo_end_date> 2002-12-12</embargo_end_date>
<use_permission accession="CC0" uri="http://creativecommons.org/publicdomain/zero/1.0/" term="Creative commons"/>
</sharing_policy>
XML Instance Representation
<...
type="xs :tokenvalue comes from list: {'closedAccess'|'embargoedAccess'|'restrictedAccess'|'openAccess'})[1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlSharingPolicy ">
<
xs:sequence >
</
xs:sequence >
<
xs:attribute name ="
type "
use ="
required ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="closedAccess "/>
<xs:enumeration value ="embargoedAccess "/>
<xs:enumeration value ="restrictedAccess "/>
<xs:enumeration value ="openAccess "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlSource
Abstract
no
Documentation
Contains information on data source. Typically specifies the source database, although the system, organization, or institution from which data is exported can also be used.
Element is re-used from the
LRG specification .
In the LSDB XML format there can be more than one source, so that information on original data providers can be included.
Example:
<source id="dmudb">
<name> DMuDB, NGRL, Manchester</name>
<url> http://www.ngrl.org.uk/Manchester/projects/informatics/dmudb</url>
<contact>
<name> Glen Dobson</name>
<address> NGRL, Genetic Medicine, St Mary's Hospital, Oxford Road, Manchester M13 9WL</address>
<phone> (+44) 0000 000 0000</phone>
<fax> (+44) 0000 000 0000</fax>
<email> example@nnn.nn.nnn</email>
</contact>
</source>
XML Instance Representation
<...
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " version="anySimpleType [0..1] " date="xs :date[0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlSource ">
<
xs:sequence >
<
xs:element name ="
name "
type ="
xs :string"/>
<
xs:element ref ="
vml :url "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :contact "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<xs:attribute name ="version "/>
<
xs:attribute name ="
date "
type ="
xs :date"/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlStrain ">
</
xs:complexType >
Super-types:
None
Sub-types:
Name
VmlSubmission
Abstract
no
Documentation
Asbtract submission element. Contains information which is common to all submission entries like lsdb and vreport elements. See the subtypes for documentation.
Schema Component Representation
<
xs:complexType name ="
VmlSubmission ">
<
xs:sequence >
<
xs:group ref ="
vml :source "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
schema_version "
type ="
xs :decimal"/>
<
xs:attribute name ="
submissionid_type "
type ="
xs :string"/>
</
xs:complexType >
Super-types:
VmlDbXRef VmlSubmitterId (by extension)
Sub-types:
None
Name
VmlSubmitterId
Abstract
no
Documentation
Randomized identifier of individual as used by submitter.
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [1] " name="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " >
</...>
Schema Component Representation
<
xs:complexType name ="
VmlSubmitterId ">
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlTissue ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlTissueDistribution
Abstract
no
Documentation
Tissue distribution
Controlled vocabulary terms for tissue disttribution:
lsdb-controlled-vocabulary-terms
constitutional
mosaic
mosaic in germline
Example:
<tissue_distribution term="mosaic"/>
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlTissueDistribution ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlUsePermission ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlVariant
Abstract
no
Documentation
Variant. Typically has the following information:
type - DNA,cDNA,RNA,AA
gene
ref_seq - reference sequence
name - name of variant, preferrably using the HGVS nomenclature
consequence - consequence (list of consequences) of the variant using for example sequence or variation ontology
pathogenicity
phenotype - known phenotypic consequence
variant_detection - variant detection method
restriction_site - site of restriction enzyme
seq_changes - effects of variant on overlapping sequence levels/features
aliases - alternative representations of the variant
<variant diploid_count="1">
<gene source="HGNC" accession="AIRE"/>
<ref_seq source="IDRefSeq" accession="D0003"/>
<name scheme="HGVS"> g.4707A>T</name>
<exon> e1</exon>
<consequence term="A point mutation in the exon 1 altering initiation codon in HSR domain "/>
<seq_changes>
<variant type="RNA">
<ref_seq source="IDRefSeq" accession="C0003"/>
<name scheme="HGVS"> r.1a>u</name>
<consequence term="predicted to change start codon to position c.268, resulting in a frameshift of translation"/>
<seq_changes>
<variant type="AA">
<ref_seq source="UniProt" accession="O43918"/>
<name scheme="HGVS"> p.Met1Leu</name>
</variant>
</seq_changes>
</variant>
</seq_changes>
</variant>
XML Instance Representation
<...
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source </...>
Schema Component Representation
<
xs:complexType name ="
VmlVariant ">
<
xs:sequence >
<
xs:element ref ="
vml :gene "
minOccurs ="
0 "/>
<
xs:group ref ="
vml :name "
minOccurs ="
0 "/>
<
xs:element ref ="
vml :panel "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :exon "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :consequence "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :frequency "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :location "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
type ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="DNA "/>
<xs:enumeration value ="cDNA "/>
<xs:enumeration value ="RNA "/>
<xs:enumeration value ="AA "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
<
xs:attribute name ="
genotypic ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="true "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
<
xs:attribute name ="
subcellular_part ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="nucleus "/>
<xs:enumeration value ="mitochondrial "/>
<xs:enumeration value ="chloroplast "/>
<xs:enumeration value ="other "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
<
xs:attribute name ="
copy_count "
type ="
xs :float"/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlVariantClass ">
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlVariantDetection
Abstract
no
Documentation
Techniques used to detect the variant.
Variant detection method. Element has attributes:
sequence template, which can be either DNA, RNA, cDNA or AA
detection technique
Optional element protocol_id (of type DbXRef) has reference to actual detection protocol.
Controlled vocabulary terms for detection techniques:
lsdb-controlled-vocabulary-terms
ARMS
CF20: CF Common Mutation Test
CF29: Analysis of 29 mutations using the Elucigene CF29 kit
CSCE: Conformation sensitive capillary electrophoresis
DGGE: Denaturing gradient gel electrophoresis
dHPLC: Denaturing high performance liquid chromatography
Heteroduplex analysis
Loss of heterozygosity analysis
Meta-PCR
MLPA: Multiplex ligation-dependent probe amplification
MS-PCR: Mutagenically separated PCR
Multiplex PCR
Not Known: The information has not been recorded or provided
Not Specified: Test information cannot be determined
PCR-PAGE
PTT: Protein Trucation Test
RNA: RNA work performed
Sequencing
SNPlex: The SNPlex™ Genotyping System from ABI
SSCP
SSCP/Heteroduplex
Example:
<variant_detection template="DNA" technique="DHPLC"/>
XML Instance Representation
<...
template="xs :tokenvalue comes from list: {'DNA'|'RNA'|'cDNA'|'AA'})[1] " technique="anySimpleType [1] " >
Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </...>
Schema Component Representation
<
xs:complexType name ="
VmlVariantDetection ">
<
xs:sequence >
</
xs:sequence >
<
xs:attribute name ="
template "
use ="
required ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="DNA "/>
<xs:enumeration value ="RNA "/>
<xs:enumeration value ="cDNA "/>
<xs:enumeration value ="AA "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
<xs:attribute name ="technique " use ="required "/>
</
xs:complexType >
Super-types:
None
Sub-types:
None
Name
VmlVariantGroup
Abstract
no
Documentation
Variant group
XML Instance Representation
<...
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " orientation="xs :tokenvalue comes from list: {'cis'|'trans'|'unknown'})[0..1] " >
Start Group: vml :variant [1..*] <
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity </...>
Schema Component Representation
<
xs:complexType name ="
VmlVariantGroup ">
<
xs:sequence >
<
xs:group ref ="
vml :variant "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :frequency "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
<
xs:attribute name ="
orientation ">
<
xs:simpleType >
<
xs:restriction base ="
xs :token">
<xs:enumeration value ="cis "/>
<xs:enumeration value ="trans "/>
<xs:enumeration value ="unknown "/>
</
xs:restriction >
</
xs:simpleType >
</
xs:attribute >
</
xs:complexType >
Super-types:
None
Sub-types:
None
XML Instance Representation
<...
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
<
vml :description>
xs :string </
vml :description>
[0..1] </...>
Schema Component Representation
<
xs:complexType name ="
VmlVariantType ">
</
xs:complexType >
Name
VmlAbstractObservationTarget
Documentation
Abstract observation target. Contains information which is common to all observation targets like individuals and panels.
id,uri - optional unqiue identifiers
original_id - optional source identifier as used in source system
organism
phenotypes -list of phenotypes
populations - list of population groups target belongs to
variants -list of variants targes has
source - original source of the information
XML Instance Representation
Start Group: vml :variant [0..*]
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant
Start Group: vml :source [0..1]
End Group: vml :source
Schema Component Representation
<
xs:group name ="
VmlAbstractObservationTarget ">
<
xs:sequence >
<
xs:element ref ="
vml :role "
minOccurs ="
0 "/>
<
xs:element ref ="
vml :phenotype "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :population "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:group ref ="
vml :variant "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:group >
Name
VmlAcknowledgement
Documentation
Acknowledgement.
Schema Component Representation
<
xs:group name ="
VmlAcknowledgement ">
<
xs:sequence >
<
xs:element name ="
name "
type ="
xs :string"/>
</
xs:sequence >
</
xs:group >
Name
VmlAnnotatable
Documentation
Abstract element which has annotations (i.e. db_xrefs and comments)
Schema Component Representation
<
xs:group name ="
VmlAnnotatable ">
<
xs:sequence >
<
xs:element ref ="
vml :db_xref "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:element ref ="
vml :comment "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:group >
Name
VmlAnnotatedObservation
Documentation
Not implemented- order of elements is not set.
Schema Component Representation
<
xs:group name ="
VmlAnnotatedObservation ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Name
VmlAnything
Documentation
Anything.
Schema Component Representation
<
xs:group name ="
VmlAnything ">
<
xs:sequence >
<xs:any minOccurs ="0 " maxOccurs ="unbounded " processContents ="skip "/>
</
xs:sequence >
</
xs:group >
Name
VmlConsequentVariants
Documentation
Variants on subsequent sequence levels
XML Instance Representation
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'cDNA'|'RNA'|'AA'})[0..1] " >
[1..*] <
vml :name
scheme="anySimpleType [0..1] " />
[1] Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
Schema Component Representation
<
xs:group name ="
VmlConsequentVariants ">
<
xs:sequence >
<
xs:element name ="
variant "
maxOccurs ="
unbounded ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
Name
VmlEvidenceOntologyTerm
Documentation
Abstract datatype (superclass) for controlled vocabulary terms related to observations. All Evidence_ontology_term elements have:
optional protocol and database crossreferences - list of protocol_id and db_xref elements
evidence codes - list of evidence_code-elements
In addition of properties inherited from ontology_class (like db_xref and comment)
<genetic_origin term="inherited">
<source term="paternal"/>
<evidence_code term="inferred"/>
</genetic_origin>
<phenotype term="Osteogenesis Imperfecta">
<protocol_id accession="observation_methodXYZ" source="protocol_database"/>
</phenotype>
<phenotype term="Autoimmune polyglandular syndrome type 1">
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
<
vml :description>
xs :string </
vml :description>
[0..1]
Schema Component Representation
<
xs:group name ="
VmlEvidenceOntologyTerm ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Name
VmlForeignElements
Documentation
Define foreign nodes for extensions.
Schema Component Representation
<
xs:group name ="
VmlForeignElements ">
<
xs:sequence >
<xs:any minOccurs ="0 " maxOccurs ="unbounded " namespace ="##other " processContents ="skip "/>
</
xs:sequence >
</
xs:group >
Name
VmlForeignNodes
Documentation
Define foreign nodes for extensions.
Schema Component Representation
<
xs:group name ="
VmlForeignNodes ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Name
VmlGenotype
Documentation
Genotype
Schema Component Representation
<
xs:group name ="
VmlGenotype ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Name
VmlLocation
Documentation
Location of variant on a reference sequence/genomic build
Example:
<location>
<ref_seq source="embl" accession="ABC1023345"/>
<start> 111111</start>
<end> 111112</end>
</location>
<location>
<ref_seq accession="build36"/>
<chr> 21</chr>
<start> 111111</start>
<end> 111112</end>
</location>
Schema Component Representation
<
xs:group name ="
VmlLocation ">
<
xs:sequence >
<
xs:element ref ="
vml :chr "
minOccurs ="
0 "/>
<
xs:element ref ="
vml :end "
minOccurs ="
0 "/>
</
xs:sequence >
</
xs:group >
Name
VmlObservation
Documentation
Abstract type. See subtypes for the documentation. Observation which has evidence code and observation protocol (dbxref).
Example:
<genetic_origin term="paternal">
<evidence_code term="inferred"/>
</genetic_origin>
<phenotype term="Osteogenesis Imperfecta">
<evidence_code term="assessed by clinician"/>
<protocol_id source="protocoldb_xyz" accession="XYZ0001"/>
</phenotype>
Schema Component Representation
<
xs:group name ="
VmlObservation ">
<
xs:sequence >
<
xs:element ref ="
vml :protocol_id "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:group >
Name
VmlObservations
Documentation
Abstract type. See subtypes for the documentation. Observation which has evidence code and observation protocol (dbxref).
Example:
<genetic_origin term="paternal">
<evidence_code term="inferred"/>
</genetic_origin>
<phenotype term="Osteogenesis Imperfecta">
<evidence_code term="assessed by clinician"/>
<protocol_id source="protocoldb_xyz" accession="XYZ0001"/>
</phenotype>
XML Instance Representation
Start Choice [0..1]
End Choice
Start Group: vml :variant [0..*]
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant
Schema Component Representation
<
xs:group name ="
VmlObservations ">
<
xs:sequence >
<
xs:choice minOccurs ="
0 ">
</
xs:choice >
<
xs:element ref ="
vml :phenotype "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
<
xs:group ref ="
vml :variant "
minOccurs ="
0 "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:group >
Name
VmlOntology
Documentation
Ontology class. Supertype for ontology terms (most of the VarioML elements are ontology or evidence ontology terms)
source - name (abbreviation) of system where term is defined e.g. SO (sequence ontology)
accession - accession number of term
term - ontology term
uri - uri of the term
dbx_ref - database cross-references
comment - comments
Example:
<phenotype term="Autoimmune polyglandular syndrome type 1" accession="240300" source="OMIM">
<db_xref accession="12345" source="pubmed"/>
<comment term="symptom"> <text> Hypoparathyroidism</text></comment>
<comment term="symptom"> <text> Addison's disease</text></comment>
<comment term="symptom"> <text> Mucocutaneous candidiasis</text></comment>
<comment term="symptom"> <text> Malabsorption</text></comment>
<comment term="symptom"> <text> Chronic active hepatitis</text></comment>
</phenotype>
XML Instance Representation
<
vml :description>
xs :string </
vml :description>
[0..1]
Schema Component Representation
<
xs:group name ="
VmlOntology ">
<
xs:sequence >
<
xs:element name ="
description "
type ="
xs :string"
minOccurs ="
0 "/>
</
xs:sequence >
</
xs:group >
Name
VmlOntologyTerm
Documentation
Abstract annotated ontology class. See supertype for more information.
XML Instance Representation
<
vml :description>
xs :string </
vml :description>
[0..1]
Schema Component Representation
<
xs:group name ="
VmlOntologyTerm ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Name
VmlRelatedVariants
Documentation
Related variants. See aliases for more information
XML Instance Representation
Start Group: vml :variant [1..*]
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
End Group: vml :variant
Schema Component Representation
<
xs:group name ="
VmlRelatedVariants ">
<
xs:sequence >
<
xs:group ref ="
vml :variant "
maxOccurs ="
unbounded "/>
</
xs:sequence >
</
xs:group >
Name
VmlSequence
Documentation
Reference and variant sequence strings. Accession number of reference sequence is given in ref_seq element (see the variant)
Example:
<sequence>
<reference> atttgatcgttc</reference>
<variant> atttgGtcgttc</variant>
</sequence>
XML Instance Representation
<
vml :variant>
xs :string </
vml :variant>
[1]
Schema Component Representation
<
xs:group name ="
VmlSequence ">
<
xs:sequence >
<
xs:element name ="
variant "
type ="
xs :string"/>
</
xs:sequence >
</
xs:group >
Name
description
Documentation
Description of gender. Used when geneder code i 9
Example:
<gender code="9">
<description term="XX-male"/>
</gender>
XML Instance Representation
<
vml :description
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :description>
Schema Component Representation
<
xs:group name ="
description ">
<
xs:sequence >
<
xs:element name ="
description ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
Model Group: factor
XML Instance Representation
<
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
Schema Component Representation
<
xs:group name ="
factor ">
<
xs:sequence >
<
xs:element name ="
pathogenicity ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
Schema Component Representation
<
xs:group name ="
genetic_source ">
<
xs:sequence >
</
xs:sequence >
</
xs:group >
Model Group: name
XML Instance Representation
<
vml :name
scheme="anySimpleType [0..1] " />
[1]
Schema Component Representation
<
xs:group name ="
name ">
<
xs:sequence >
<
xs:element name ="
name ">
<
xs:complexType mixed ="
true ">
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
XML Instance Representation
<
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
Schema Component Representation
<
xs:group name ="
pathogenicity ">
<
xs:sequence >
<
xs:element name ="
pathogenicity ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
Model Group: source
Schema Component Representation
<
xs:group name ="
source ">
<
xs:sequence >
<
xs:element name ="
source ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
XML Instance Representation
<
vml :variant
id="vml :VmlId [0..1] " uri="vml :VmlUri [0..1] " type="xs :tokenvalue comes from list: {'DNA'|'cDNA'|'RNA'|'AA'})[0..1] " genotypic="xs :tokenvalue comes from list: {'true'})[0..1] " subcellular_part="xs :tokenvalue comes from list: {'nucleus'|'mitochondrial'|'chloroplast'|'other'})[0..1] " copy_count="xs :float[0..1] " >
[1] Start Group: vml :name [0..1] <
vml :name
scheme="anySimpleType [0..1] " />
[1] End Group: vml :name Start Group: vml :pathogenicity [0..*] <
vml :pathogenicity
scope="xs :tokenvalue comes from list: {'individual'|'family'|'population'})[0..1] " panel_ref="vml :VmlId [0..1] " source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] Start Group: vml :factor [0..*] <
vml :pathogenicity
source="anySimpleType [0..1] " accession="anySimpleType [0..1] " uri="vml :VmlUri [0..1] " term="anySimpleType [1] " >
[1] <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :factor <
vml :description>
xs :string </
vml :description>
[0..1] Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :pathogenicity>
End Group: vml :pathogenicity Start Group: vml :source [0..1] End Group: vml :source Start Group: vml :VmlForeignElements [0..*] Allow any elements from a namespace other than this schema's namespace (skip validation). [0..*]
End Group: vml :VmlForeignElements </
vml :variant>
Schema Component Representation
<
xs:group name ="
variant ">
<
xs:sequence >
<
xs:element name ="
variant ">
<
xs:complexType >
<
xs:complexContent >
</
xs:complexContent >
</
xs:complexType >
</
xs:element >
</
xs:sequence >
</
xs:group >
Super-types:
xs :stringVmlAccession (by restriction)
Sub-types:
None
Name
VmlAccession
Content
Documentation
Accession number of the term
Schema Component Representation
<
xs:simpleType name ="
VmlAccession ">
<
xs:restriction base ="
xs :string"/>
</
xs:simpleType >
Super-types:
VmlDate VmlCreationDate (by restriction)
Sub-types:
None
Name
VmlCreationDate
Content
Union of following types:
Documentation
Date when submitted information was created
Schema Component Representation
<
xs:simpleType name ="
VmlCreationDate ">
</
xs:simpleType >
Super-types:
None
Sub-types:
Name
VmlDate
Content
Union of following types:
Documentation
Date can include time, time zone, year, and month.
Schema Component Representation
<
xs:simpleType name ="
VmlDate ">
<
xs:union memberTypes ="
xs :gYear xs :gYearMonth xs :date xs :dateTime"/>
</
xs:simpleType >
Super-types:
VmlDate VmlDobDate (by restriction)
Sub-types:
None
Name
VmlDobDate
Content
Union of following types:
Documentation
Date when submitted information was modified.
Example:
<modification_date> 2001</modification_date>
<modification_date> 2001-09</modification_date>
<modification_date> 2001-09-10</modification_date>
Schema Component Representation
<
xs:simpleType name ="
VmlDobDate ">
</
xs:simpleType >
Simple Type: VmlId
Super-types:
xs :stringVmlId (by restriction)
Sub-types:
None
Name
VmlId
Content
Documentation
Unique identifier which applies to various elements and contexts in the LSDB schema.
Can be local or assigned by a consortium.
Schema Component Representation
<
xs:simpleType name ="
VmlId ">
<
xs:restriction base ="
xs :string"/>
</
xs:simpleType >
Super-types:
VmlDate VmlModificationDate (by restriction)
Sub-types:
None
Name
VmlModificationDate
Content
Union of following types:
Documentation
Date when submitted information was modified.
Example:
<modification_date> 2001</modification_date>
<modification_date> 2001-09</modification_date>
<modification_date> 2001-09-10</modification_date>
Schema Component Representation
<
xs:simpleType name ="
VmlModificationDate ">
</
xs:simpleType >
Super-types:
xs :stringVmlNameStr (by restriction)
Sub-types:
None
Name
VmlNameStr
Content
Documentation
Name of source.
Schema Component Representation
<
xs:simpleType name ="
VmlNameStr ">
<
xs:restriction base ="
xs :string"/>
</
xs:simpleType >
Simple Type: VmlUri
Super-types:
xs :anyURIVmlUri (by restriction)
Sub-types:
None
Name
VmlUri
Content
Documentation
URI for various elements and contexts in the LSDB schema.
In context of ontology term:
Note that the uri should be used only for resolving the ontology term, not for the content of implementing element (e.g. phenotype).
Schema Component Representation
<
xs:simpleType name ="
VmlUri ">
<
xs:restriction base ="
xs :anyURI"/>
</
xs:simpleType >
Simple Type: Vmlurl
Super-types:
xs :anyURIVmlurl (by restriction)
Sub-types:
None
Name
Vmlurl
Content
Documentation
URL for various elements and contexts in the LSDB schema.
Schema Component Representation
<
xs:simpleType name ="
Vmlurl ">
<
xs:restriction base ="
xs :anyURI"/>
</
xs:simpleType >
Complex Type:
Schema Component Type
AusAddress
Schema Component Name
Super-types:
Address < AusAddress (by extension)
Sub-types:
QLDAddress (by restriction)
If this schema component is a type definition, its type hierarchy is shown in a gray-bordered box.
The table above displays the properties of this schema component.
XML Instance Representation
<... country="Australia " >
<unitNo> string </unitNo> [0..1] <houseNo> string </houseNo> [1] <street> string </street> [1] Start Choice [1] <city> string </city> [1] <town> string </town> [1] End Choice <state> AusStates </state> [1] <postcode> string <<pattern = [1-9][0-9]{3}>> </postcode> [1] ? </...>
The XML Instance Representation table above shows the schema component's content as an XML instance.
The minimum and maximum occurrence of elements and attributes are provided in square brackets, e.g. [0..1].
Model group information are shown in gray, e.g. Start Choice ... End Choice.
For type derivations, the elements and attributes that have been added to or changed from the base type's content are shown in bold .
If an element/attribute has a fixed value, the fixed value is shown in green, e.g. country="Australia".
Otherwise, the type of the element/attribute is displayed.
If the element/attribute's type is in the schema, a link is provided to it. For local simple type definitions, the constraints are displayed in angle brackets, e.g. <<pattern = [1-9][0-9]{3}>>.
If a local element/attribute has documentation, it will be displayed in a window that pops up when the question mark inside the attribute or next to the element is clicked, e.g. <postcode>.
Schema Component Representation
<complexType name ="AusAddress ">
<complexContent >
<extension base ="Address
<sequence >
<element name ="state " type ="AusStates
<element name ="postcode ">
<simpleType >
<restriction base ="string
<pattern value ="[1-9][0-9]{3} "/>
</restriction >
</simpleType >
</element >
</sequence >
<attribute name ="country " type ="string fixed ="Australia "/>
</extension >
</complexContent >
</complexType >
The Schema Component Representation table above displays the underlying XML representation of the schema component. (Annotations are not shown.)
Abstract
All Model Group in any order in instances. See: http://www.w3.org/TR/xmlschema-1/#element-all .
Choice Model Group Only one from the list of child elements and model groups can be provided in instances. See: http://www.w3.org/TR/xmlschema-1/#element-choice .
Collapse Whitespace Policy
Disallowed Substitutions substitution is specified, then substitution group members cannot be used in place of the given element declaration to validate element instances. If derivation methods , e.g. extension, restriction, are specified, then the given element declaration will not validate element instances that have types derived from the element declaration's type using the specified derivation methods. Normally, element instances can override their declaration's type by specifying an xsi:type attribute.
Key Constraint Uniqueness Constraint , but additionally requires that the specified value(s) must be provided. See: http://www.w3.org/TR/xmlschema-1/#cIdentity-constraint_Definitions .
Key Reference Constraint Key Constraint or Uniqueness Constraint . See: http://www.w3.org/TR/xmlschema-1/#cIdentity-constraint_Definitions .
Model Group http://www.w3.org/TR/xmlschema-1/#Model_Groups .
Nillable xsi:nil attribute. The xsi:nil attribute is the boolean attribute, nil , from the http://www.w3.org/2001/XMLSchema-instance namespace. If an element instance has an xsi:nil attribute set to true, it can be left empty, even though its element declaration may have required content.
Notation http://www.w3.org/TR/xmlschema-1/#cNotation_Declarations .
Preserve Whitespace Policy
Prohibited Derivations
Prohibited Substitutions
Replace Whitespace Policy
Sequence Model Group in the specified order in instances. See: http://www.w3.org/TR/xmlschema-1/#element-sequence .
Substitution Group members of a substitution group can be used wherever the head element of the substitution group is referenced.
Substitution Group Exclusions
Target Namespace
Uniqueness Constraint http://www.w3.org/TR/xmlschema-1/#cIdentity-constraint_Definitions .
Classes and properties from other ontologies can be used
together with VarioML.
This section lists the most important external classes and
properties that can be used with VarioML in a meaningful way. This
list is not and cannot be exhaustive because many RDF ontologies
can be used together.
These are the ontology namespaces referenced:
We would like to acknowledge the contributions of ..
We would also like to acknowledge the many helpful suggestions
from members of Gen2Phen , and we
thank ..
Sharing
data
between LSDBs and central repositories. Hum Mutat, 2009. 30(4): p.
493-5.Recommendations for
locus-specific databases and their curation.
Hum Mutat. 2008 Jan;29(1):2-5.Swertz, M.A., et al., Molecular Genetics Information System
(MOLGENIS): alternatives in developing local experimental genomics
databases. Bioinformatics, 2004. 20(13): p. 2075- 83.
Swertz, M.A. and R.C. Jansen, Beyond standardization: dynamic
software infrastructures for systems biology. Nat Rev Genet, 2007.
8(3): p. 235-43.
http://www.ietf.org/rfc/rfc2119.txt .http://www.ietf.org/rfc/rfc2396.txt .
4.5.2010: phenotype_refinement added into variant
4.5.2010: mixed content removed ontology types. Mandatory
attribute term is used instead of text node (as in FuGE).
4.5.2010: optional desc-element added into ontology types for
free form textual descriptions
4.5.2010: mixed content remove from comment. comment text goes
into new "text" sub-element.
4.5.2010: mixed content removed from name. it do not have
comment, evidence and dbxref subelements any more
4.5.2010: mixed content removed from exon. it do not have
comment, evidence and dbxref subelements any more
4.5.2010: TBD: population: ethnic, region (inc. nation),
language, other
4.5.2010: acknowledgement added into source. See AIREbase
example
4.5.2010: contact: added phone and fax
4.5.2010: source: added version and date attributes (see
AIREbase example)
4.5.2010: lsdb:created time stamp
4.5.2010: ethnicity -> population (see below)
4.5.2010: geographical_location -> population (see
below)
4.5.2010: population has type (for
"region","ethnicity","race"...). type is AnnotatedOntology
4.5.2010: individual: location of gender and original_id
swapped
contact1 contact2